

LatPack is a collection of tools related to the folding simulations of lattice-protein models with arbitrary energy function. It is therefore embedded in our research field Simplified Protein Models.
./configuremakemake installYou can download the LatPack source code including configure scripts:
LatFit is a tool allowing for the creation of high-resolution models of full-atom protein structures in lattice models. It enables the fitting of backbone and side chain models of a given protein in PDB format.
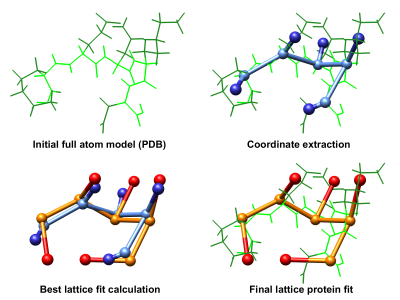
We follow a chain elongation procedure similar to that described by Park and Levitt (JMB, 1995). Residues are placed on the lattice sequentially starting from the amino terminus. For each residue placement, the current best lattice fit is iteratively extended. Each extension is evaluated via RMSD, and the best fit further extended until full chain length is achieved.
The coordinates fitted are user-defined. By default, LatFit takes the C_alpha atom coordinates for the backbone and the center of mass of non-hydrogen side chain atoms as the side chain.
LatFit is embedded in our research field Simplified Protein Models.
./configuremakemake install