Libraries
Overview

This C++ programming library provides classes and functions often used in
the bioinformatics context of our group. It collects data structures and
class hierarchies for biomolecule modeling and general useful
functionalities as program parameter parsing, string conversions, etc.
Current Status
- String conversions etc.
- Program parameter parser
- Random number generation
- Lattice proteins and related operations (e.g. move sets)
- RNA support
- …
For a full list of features, see documentation.
Documentation
Downloads
You can download the BIU-library including configure scripts:
Contributing Group Members
- Martin Mann
- Sebastian Will
- Daniel Maticzka
- Hannes Kochniß
Energy Landscape Library - ELL

The ELL-library provides a platform for generic algorithms to study kinetics
and structure of energy landscapes with discrete states. These algorithms
need an abstract representation of these states to be applied to a multitude
of state instances and their corresponding energy or fitness landscapes.
Main Publications
Current Status
Generic Algorithms
- A set of walks to traverse energy landscapes (random, adaptive, gradient, Monte Carlo)
- Exhaustive landscape flooding (single and multiple basins)
- Sampling-based macro-state transition rate estimation
Landscape Models
- Lattice protein representations with Pull-Move and Pivot-Move neighborhoods for different lattice models with arbitrary distance-based energy functions
- RNA representation with single base pair indel, interfacing the Vienna RNA package
- Ising spin glass models (ring of spins)
- Number Partitioning Problem (ring of spins)
General Features
- Strict separation of algorithm and landscape model layer
- Memory-efficient compressed State representations
- Generic neighborhood generators
- Global and flexible setup of random number generators
For a full list of features, see documentation.
Documentation
Downloads
You can download the ELL-library including configure scripts and documentation:
For installation instructions, see documentation and the included INSTALL file.
Dependencies
The ELL depends on the following libraries:
Contributing Group Members
- Martin Mann
- Sebastian Will
- Daniel Maticzka
- Hannes Kochniß
- Andreas Richter
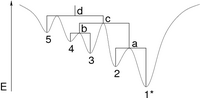
Based on the energy landscape library, a set of tools to investigate the
structure and topology of RNA energy landscapes has been implemented. These
programs allow for:
- Minima sampling
- Exact calculation of barrier trees or saddle networks
- Approximation of barrier trees
- Barrier estimation using a heuristic by Morgan and Higgs
Downloads
You can download the RNA energy landscape tools including configure scripts and documentation:
For installation instructions, see the included INSTALL file.
Dependencies
The RNA energy landscape tools depend on the following libraries:
Contributing Group Members
- Martin Mann
- Andreas Richter
- Sebastian Will
- Hannes Kochniß
- Daniel Maticzka
- Dmitry Gubenko




