

Constraints are an extremely successful technology that has a vast application area throughout informatics. Due to our expertise in both areas, constraint technology and bioinformatics, we are especially interested in the application of constraints to biological problems.
In the area of Bioinformatics, constraints already were successfully applied to problems related to:
All these problems can be naturally formalized using constraints over finite domains or intervals of reals.
Recently, we showed how the fundamental bioinformatics problem of sequence alignment can be solved by modern inference-based constraint methods. The in-depth study of constrained-based protein structure prediction by our group promoted the development of new search strategies and, in particular, general symmetry breaking.
In this area, we investigate the application of constraint techniques to the alignment problem. Our focus is to allow for a more declarative form in order to incorporate biological knowledge.
Recently, we succeeded in constructing a constraint-model for sequence alignment that can be efficiently evaluated using the inference-based constraint solving strategy of Cluster Tree Elimination. At the same time, the model can be extended by additional constraints (in order to incorporate biological prior knowledge) and is still efficiently solved.
Hitherto, our work results in a general framework for constrained alignment where almost arbitrary constraints can be imposed and even combined arbitrarily. Possible constraints range from all the constraints that were discussed for sequence alignment before up to complex structural constraints.
A web server for constrained alignment is in preparation.
In constraint programming as well as in bioinformatics, solving search problems that contain lots of symmetries is a frequent task. (For example, in protein folding, symmetries are rotations, reflections, and translation of a conformation). These symmetries blow up the search tree and search times. We first encountered the problem of breaking the symmetries in protein structure prediction. In this project, we are interested in general schemes for symmetry breaking and identifying interesting sub-classes of symmetries. Furthermore, we are interested in applying symmetry breaking to problems of bioinformatics.
We have developed the first general method for breaking arbitrary symmetries. This method is based on dynamic insertion of symmetry breaking constraints during constraint-based search.
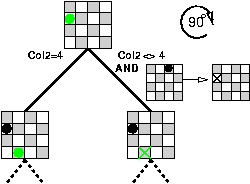
Besides handling geometrical symmetries and permutation symmetries, we identified the subclass of partial symmetries and showed how the approach can break them too.
The counting of solutions of a given Constraint Satisfaction Problem (CSP) is much more complex than deciding on satisfiability and has gained much interest recently. The solving of a CSP is, in general, NP-complete, whereby counting all solutions is even harder and in the complexity class #P.
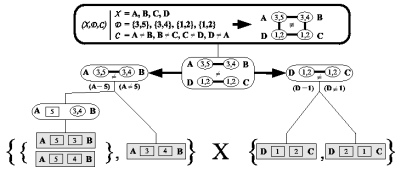
We have integrated AND/OR-search into a full-featured constraint programming system utilizing the benefits of global constraints and dynamic search heuristics. It was implemented using the free constraint programming library Gecode and will be part of the official release.